Class Imbalance comes in Like a Lion
All you need to know to handle Class Imbalance.
In a world without class imbalance we might've been heroes.
- Neural Networks
Keeping aside the fact that I butchered one of the greatest Video Game quotes of all time class imbalance can be a tricky thing to handle especially if you are a beginner. When I first encountered class imbalance I treated it normally, I know right, and not just that I measured the accuracy to judge the performance. Needless to say, that went quite badly, and to avoid this happening to you let me help out in avoiding an embarrassing situation in front of your teacher or whoever you report to.
Class Imbalance refers to a condition where the no. of data points corresponding to a class overpowers the other in a significant way. This could happen cause of bias towards a particular class during data collection, error during labeling, etc. I mean the cause doesn't matter once the data is served so all you can do now is see what you can do with whatever you have.
We'll see how to tackle class imbalance in different domains like structured data, NLP, and CV. We'll see some of the techniques you can use to modify your data to balance out the class ratio and we'll talk about how you can fix this thing on a model level without modifying the data itself.

Loading Our Data
I believe that the correct way to learn a concept is by applying what you learn in theory and that's why I'll be putting code for you to see how we are actually going to apply what we are talking about. For the purpose of this article I've decided to use the classic dataset used to teach class imbalance i.e. Credit Card Fraud Detection.
df = pd.read_csv('/kaggle/input/creditcardfraud/creditcard.csv')
X = df.drop('Class', axis = 1)
Y = df['Class']
Y.value_counts()
Output:-
0 284315
1 492
Name: Class, dtype: int64
I guess it's safe to say that our data is messed up. YAY! So now that we have our data let's get some action.
Choosing The Correct Metric
First things first, whenever you see class imbalance you have to ditch Accuracy then and there no questions asked. Think about it you give your model data to tell if a patient is diabetic or not but the data only has 10% samples for diabetic entries, which means the model can attain 90% accuracy just by predicting not diabetic every time. What you wanna see is how well the model can classify the diabetic entries or our minority class. For this, we can use various metrics:-
- Precision: Out of all entries classified as class A how many were correctly classified.
- Recall: How many entries of class A was our model able to recall correctly.
- F1-Score: Harmonic mean of Precision and Recall.
- ROC-AUC Score: Area under Curve of plot between Specificity and Sensitivity Values at different thresholds.
- PR Curve: Plot between Precision and Recall Values at different thresholds.
MCC and Kappa Score
So you get the gist right, Accuracy is not always accurate. But apart from the above-mentioned, I wanna talk about one more matrix. The Dark Horse of the Evaluation Metrics and arguably the best classification metric Matthew's Coherent Coefficient or MCC Score if you may.
The above metrics are fine too but MCC Score is much more reliable since it gives a good score only when all the portions of the confusion matrix give good results i.e. TP, FP, TN, and FN.
MCC is designed for binary classification but it can be used for multi-class classification using micro or macro averaging. We also have Kappa Score that can be used for both imbalanced and multi-class data. Note that there are many papers that argue the reliability of Kappa score and many papers that defend it. Mostly revolving around its unwanted behavior but let's leave it a topic for another blog.
Scoring The Baseline Results
Well, I hope you were able to grasp the importance of proper metrics when dealing with an imbalanced dataset. So let's start by checking the performance of our baseline model. Now in order to score our model, we'll have to split the data into training and testing splits but our data is imbalanced so we can't just do random splits. We need the data to retain the original class ratio and for that, we have the stratify parameter in train_test_split itself.
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(X, Y,
test_size = 0.25,
random_state = 1,
stratify = Y)
Well that was easy, wasn't it? Now let's go ahead and train our baseline model and check its baseline metrics using classification_report and confusion_matrix.
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report, confusion_matrix
clf = RandomForestClassifier()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
print(f'Confusion Matrix:-\n {confusion_matrix(y_test,y_pred)}\n')
print(classification_report(y_test, y_pred))
Output:-
Confusion Matrix:-
[[71073 6]
[ 17 106]]
precision recall f1-score support
0 1.00 1.00 1.00 71079
1 0.95 0.86 0.90 123
accuracy 1.00 71202
macro avg 0.97 0.93 0.95 71202
weighted avg 1.00 1.00 1.00 71202
Choosing Suitable Algorithm?
Now that we understand the importance of metrics in measuring the performance of a model in class imbalance, we can move on and check if there are any algorithms that aren't really bothered by class imbalance.
I mean on paper KNN shouldn't be bothered with class imbalance but there is something Hellinger Distance Decision Trees, basically decision trees that use Hellinger Distance as the split criterion. They were created to tackle the effect of imbalance on decision trees.
There is also a way by which you can modify your algorithm to give importance to minority class prediction by the use of class weights. Let's talk more about this cause why not.
Cost-Sensitive Algorithms
Please don't be intimidated by the name it's a rather simple concept, basically you assign weights to each class what these signify is that how much will the algorithm be penalized for a misclassification for an entry of a class. There are many algorithms in sklearn that support class weighing and few that don't support it.
The weights are assigned to the class such that the minority class has a higher weight than the majority class. We would expect that the algorithm trained on class weights will perform better as compared to the standard one.
In order to pass weights to the algorithm, you can simply pass the dictionary with key as class and value as the weight to the corresponding key to the class_weight parameter. Let's try doing this in our classifier.
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report, confusion_matrix
clf = RandomForestClassifier(verbose = 100, class_weight = {0:600,1:1})
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
print(f'Confusion Matrix:-\n {confusion_matrix(y_test,y_pred)}\n')
print(classification_report(y_test, y_pred))
Output:-
precision recall f1-score support
0 1.00 1.00 1.00 71079
1 0.92 0.88 0.90 123
accuracy 1.00 71202
macro avg 0.96 0.94 0.95 71202
weighted avg 1.00 1.00 1.00 71202
Recall value seems to have increased a bit that means our cost-sensitive model is able to recall more values from minority class in the testing set. You can try out different combinations to check if you can get better results.
Now one question that may arise in your mind is, what weights should I assign to which class? There is a simple answer to this question i.e. by tuning the weights. You can select a range of values and using GridSearch to find which ones work the best.

Let's say you are a daredevil and wanna tune the weights then, you can try using compute_class_weight() utility in sklearn to compute class weights and use them as the weights for the algorithm. In my experience, it rarely gives the best result as compared to tuned ones. But tuning is actually pretty simple, below I'll tell you how to do it and I want you to try it out by redefining the param_grid according to what you think is the best. You can comment on your findings if you like too 😃
from sklearn.model_selection import GridSearchCV
param_grid = {
'class_weight' : [{0:600,1:1}, {0:700,1:1}, {0:800,1:1}]
}
grid = GridSearchCV(RandomForestClassifier(), param_grid = param_grid)
grid.fit(x_train, y_train)
y_pred = clf.predict(x_test)
print(f'Confusion Matrix:-\n {confusion_matrix(y_test,y_pred)}\n')
print(classification_report(y_test, y_pred))
In neural networks to you can train your model with assigned class weights to tackle the issue of class imbalance. The syntax is pretty similar in the sense you just pass the class weights to the network. In Tensorflow you pass weights in the fit() function and in PyTorch you pass weights in the Loss function.
# PyTorch - Pass weight tensor in loss function
pytorch_weights = torch.tensor([0.99, 0.1])
criterion = nn.NLLLoss(weight = pytorch_weights)
# TensorFlow - Pass weight dictionary in fit function
tf_weights = { 0 : 99,
1 : 1 }
model.fit(x_train, y_train, batch_size = 50, class_weight = tf_weights)
Handling Class Imbalance with Data Modification
No no I'm not talking about modifying the values of the entries but I'm talking about how we can remove or add entries corresponding to a class to the existing data in order to balance the class ratio. There are quite a few ways to go about it and we'll explore almost all of them in-depth. Once you have balanced out the classes it basically you good old classification problem. The ones that we'll learn about are:-
- Undersampling - Fix for the Lazy
- Oversampling - Jugaad
- SMOTE - Fix from Logic
- ADASYN - SMOTE's Sibling
It'll be better If you install imblearn library since that's what we'll be using to implement the above. You can install it via the following command:-
pip install imblearn
Undersampling - Fix for the Lazy
Random Undersampling is a way to balance class by removing entries from the majority class randomly. I know right makes no sense, you are basically randomly deleting entries from the majority class. Not many people are fond of this mainly cuz it leads to loss of information, I too think it's stupid to lose that precious labeled data.

from imblearn.under_sampling import RandomUnderSampler
us = RandomUnderSampler()
x_train, y_train = us.fit_resample(x_train, y_train)
clf = RandomForestClassifier()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
print(f'Confusion Matrix:-\n {confusion_matrix(y_test,y_pred)}\n')
print(classification_report(y_test, y_pred))
Output:-
Confusion Matrix:-
[[68666 2413]
[ 8 115]]
precision recall f1-score support
0 1.00 0.97 0.98 71079
1 0.05 0.93 0.09 123
accuracy 0.97 71202
macro avg 0.52 0.95 0.53 71202
weighted avg 1.00 0.97 0.98 71202
But, what if we undersample our data smartly? There are ways and algorithms that can be used to removed redundant data and reduce the issue of losing important information. Not just but sometimes a combination of both oversampling and undersampling can achieve good results too. But enough with that let's learn about those smart undersampling methods.
Near Miss Undersampling
This type of undersampling basically determines the samples from the majority class that should be retained based on the distance between that sample and minority class samples. It has 3 variations to it:-
- Version - 1: Keeps the ones that have the minimum average distance from the nearest three minority class samples.
- Version - 2: Keeps the ones that have the minimum average distance from the farthest three minority class samples.
- Version - 3: Keeps the ones that have the minimum average distance from all minority class samples.
from imblearn.under_sampling import NearMiss
nm_us = NearMiss(version = 3)
x_train, y_train = nm_us.fit_resample(x_train, y_train)
clf = RandomForestClassifier()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
print(f'Confusion Matrix:-\n {confusion_matrix(y_test,y_pred)}\n')
print(classification_report(y_test, y_pred))
Output:-
Confusion Matrix:-
[[68681 2398]
[ 8 115]]
precision recall f1-score support
0 1.00 0.97 0.98 71079
1 0.05 0.93 0.09 123
accuracy 0.97 71202
macro avg 0.52 0.95 0.54 71202
weighted avg 1.00 0.97 0.98 71202
I've tried quite a few examples and in most case version 3 really works better as compared to the other 2.
Tomek Links Undersampling
This type of undersampling has a simple yet neat approach it focuses on removing the majority sample of the Tomek link. Tomek link is defined as the points that are closest to each other and both belonging to different classes, kinda like Romeo and Juliet except here only one of them dies, an apology to all R&J fans. Anyways let's try our hand on Tomek links.
from imblearn.under_sampling import TomekLinks
tomek_us = TomekLinks()
x_train, y_train = tomek_us.fit_resample(x_train, y_train)
clf = RandomForestClassifier(verbose = 100)
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
print(f'Confusion Matrix:-\n {confusion_matrix(y_test,y_pred)}\n')
print(classification_report(y_test, y_pred))
Here since only Tomek links are deleted the class balance isn't completely achieved rather only ambiguous points are removed.
Random Oversampling - Jugaad
This is the exact opposite of what we did in undersampling instead of removing points from the majority class we explode the minority class by filling data with samples from the minority class chosen at random with repetition and hence achieving class balance. There are ways to explode minority class samples smartly and logically like by generating synthetic data but we'll talk about them shortly.
from imblearn.over_sampling import RandomOverSampler
os = RandomOverSampler()
x_train, y_train = os.fit_resample(x_train, y_train)
clf = RandomForestClassifier(verbose = 100)
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
print(f'Confusion Matrix:-\n {confusion_matrix(y_test,y_pred)}\n')
print(classification_report(y_test, y_pred))
Output:-
Confusion Matrix:-
[[71072 7]
[ 17 106]]
precision recall f1-score support
0 1.00 1.00 1.00 71079
1 0.94 0.86 0.90 123
accuracy 1.00 71202
macro avg 0.97 0.93 0.95 71202
weighted avg 1.00 1.00 1.00 71202
SMOTE - Fix from Logic
SMOTE, or Synthetic Minority Oversampling TEchnique, is a way through which we oversample the data by generating synthetic data based on the provided samples of the minority classes. The steps required to create synthetic data are actually quite simple:-
- Take samples from the minority class
- Join these samples via lines
- Pick points that lie on these lines at random until you achieve class balance
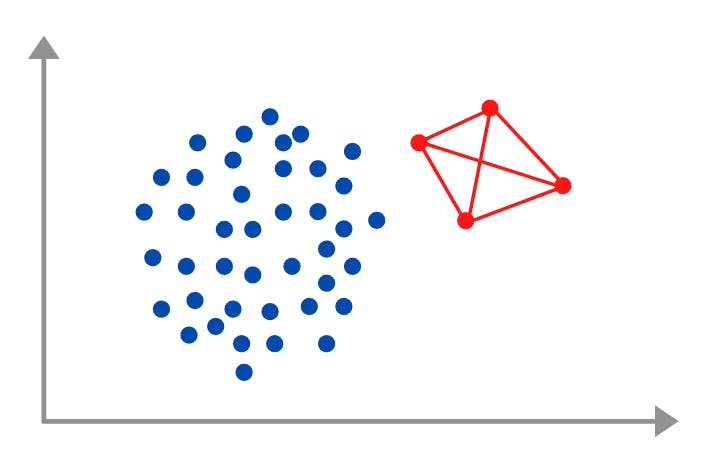
Take the above picture, which took me 30 mins to make, as an example, we only have 4 red points in the minority sample but if we join those points and add points that lie on the red line to the existing dataset then we can balance the class ratio. This basically how SMOTE works. Simple right!
from imblearn.over_sampling import SMOTE
smote = SMOTE()
x_train, y_train = smote.fit_resample(x_train, y_train)
clf = RandomForestClassifier(verbose = 100)
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
print(f'Confusion Matrix:-\n {confusion_matrix(y_test,y_pred)}\n')
print(classification_report(y_test, y_pred))
Output:-
Confusion Matrix:-
[[71066 13]
[ 14 109]]
precision recall f1-score support
0 1.00 1.00 1.00 71079
1 0.89 0.89 0.89 123
accuracy 1.00 71202
macro avg 0.95 0.94 0.94 71202
weighted avg 1.00 1.00 1.00 71202
Hmm, that seems fine recall went up a bit but precision took a hit. There are other variates to SMOTE that you can explore and learn about too. But for now, it's time to move on to the next method.
ADASYN - SMOTE's Sibling
ADASYN, or Adaptive Synthetic, is a way through which we oversample the data by generating synthetic data based on the density of majority neighbors around a minority sample. The difference between SMOTE and ADASYN is that in ADASYN the no. of samples generated around a point depends on the density distribution r_x of that point whereas in SMOTE all minority samples have equal weights.
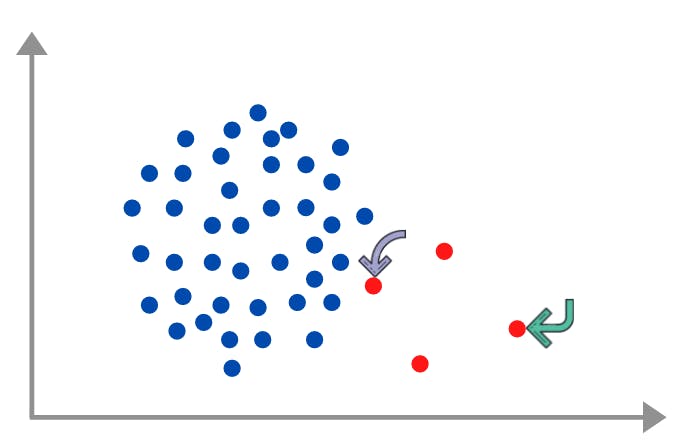
For example in the above image purple point will have more samples generated around it rather than the green arrow one. Let's understand the steps required in ADASYN:-
Calculate the no. of points to be generated, denoted by G. Here beta is the balance factor which if kept 1 generates a point to achieve perfect class balance. $$ G = ( n_M - n_m) * {\beta} $$ where n_M <- No. of majority class samples and n_m <- No. of minority class samples
For all i that belong to minority class we find the ratio:- $$ r_i = Δ_i / K $$ where K is the no. of neighbors and Δ represents the no. of samples belonging to the majority class out of those K neighbors.
Convert the above to probability distribution. $$ \hat{r_i} = r_i / \sum{r_i} $$
Calculate the no. of data points to be generated around each minority sample. Then for each minority sample, you generate gᵢ no. of samples. $$ g_i = \hat{r_i} * G $$
from imblearn.over_sampling import ADASYN
adasyn = ADASYN()
x_train, y_train = adasyn.fit_resample(x_train, y_train)
clf = RandomForestClassifier(verbose = 100)
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
print(f'Confusion Matrix:-\n {confusion_matrix(y_test,y_pred)}\n')
print(classification_report(y_test, y_pred))
Output:-
Confusion Matrix:-
[[71065 14]
[ 14 109]]
precision recall f1-score support
0 1.00 1.00 1.00 71079
1 0.89 0.89 0.89 123
accuracy 1.00 71202
macro avg 0.94 0.94 0.94 71202
weighted avg 1.00 1.00 1.00 71202
Handling Class Imbalance in Image Classification
In Image Classification, the usual way to handle class imbalance is to explode the data using Oversampling or via Data Augmentation. In oversampling, it's pretty simple you copy and you replicate like the good old way.
In Oversampling by Data Augmentation, you can generate samples by changing the nature of the image itself. For those who don't know Image Augmentation refers to changing the aspects of an image like its scale, angle orientation, etc. Keep in mind I'm not talking about just applying augmentation to the image but saving those augmentations if you do the first the dataset will be still imbalanced.
Another way to tackle this is by passing class_weights to the CNN model. Now unless you are a person who loves pain chances are you'll be using a CNN model or a pre-trained model for image classification for which we can define class_weights and train the network accordingly.
Handling Class Imbalance in Text Classification
Text Classification for the imbalanced set is also similar, we can use class_weights to define penalty for misclassification of each class. Chances are you might be using LSTM, BERT, etc. for which we can utilize class_weights.
We can also remove duplicate sentences too. Like if you have 2 sentences, "Bag is in the room" and "Bag is in room", welp they are basically the same so we can remove one of them. Removing such duplicate messages will help you reduce the size of your majority class.
The next way is oversampling the google old random way i.e. Random Oversampling or you can explode minority samples with text augmentations. Wait what? Text Augmentations!? I mean in images we rotate, scale up, crop, rotate, etc. but what can we do with text? In text augmentation we start by tokenizing sentences then we can shuffle and rejoin them to generate new texts. We can also replace adjectives, verbs, etc. by its a synonym to generate text with the same meaning.
There is another way where you are converting English text to a language and converting back to English using language translation.
From Me to You...
Boy was that a lot of information to grasp. Imbalance classification can be a pain I mean in a perfect world there would be no imbalance, sadly we live in a world where 4-koma mangas rarely get an anime adaptation, so unfair. The point is where is a will there is a way and I hope I was able to guide you through that way properly. See you in the next article!
